Publications
Chronological publications in which Ze Zhang was involved.
2025
-
 一种高含水量样本的空间转录组制备方法及其应用张泽 , 冯春露 , 庄镇堃 , 陈成 , 胡馨月 , and 陈铎元CN Patent, Nov 2025
一种高含水量样本的空间转录组制备方法及其应用张泽 , 冯春露 , 庄镇堃 , 陈成 , 胡馨月 , and 陈铎元CN Patent, Nov 2025本发明公开了一种高含水量样本的空间转录组制备方法及其应用。所述制备方法包括以下步骤:对高含水量生物样本依次进行固定液固定、明胶置换、包埋、切片处理,得到切片样本。本发明高含水量生物的空间转录组样本的制备方法通过优化组织处理、包埋和切片流程,确保切片形态完整性和RNA完整性,满足空间转录组测序的要求,且本发明的制备方法具有广泛的适用性,为空间转录组测序在高含水量样本中的应用提供了可靠的技术支持。
@article{CN120275136B, author = {张泽 and 冯春露 and 庄镇堃 and 陈成 and 胡馨月 and 陈铎元}, title = {一种高含水量样本的空间转录组制备方法及其应用}, journal = {CN Patent}, year = {2025}, month = nov, day = {04}, pages = {}, volume = {G01N1/28; G01N1/30; G01N1/36; G01N1/42; G01N1/34; C12Q1/6806; C12Q1/6869; G01N21/64}, number = {}, doi = {}, url = {}, eprint = {}, } -
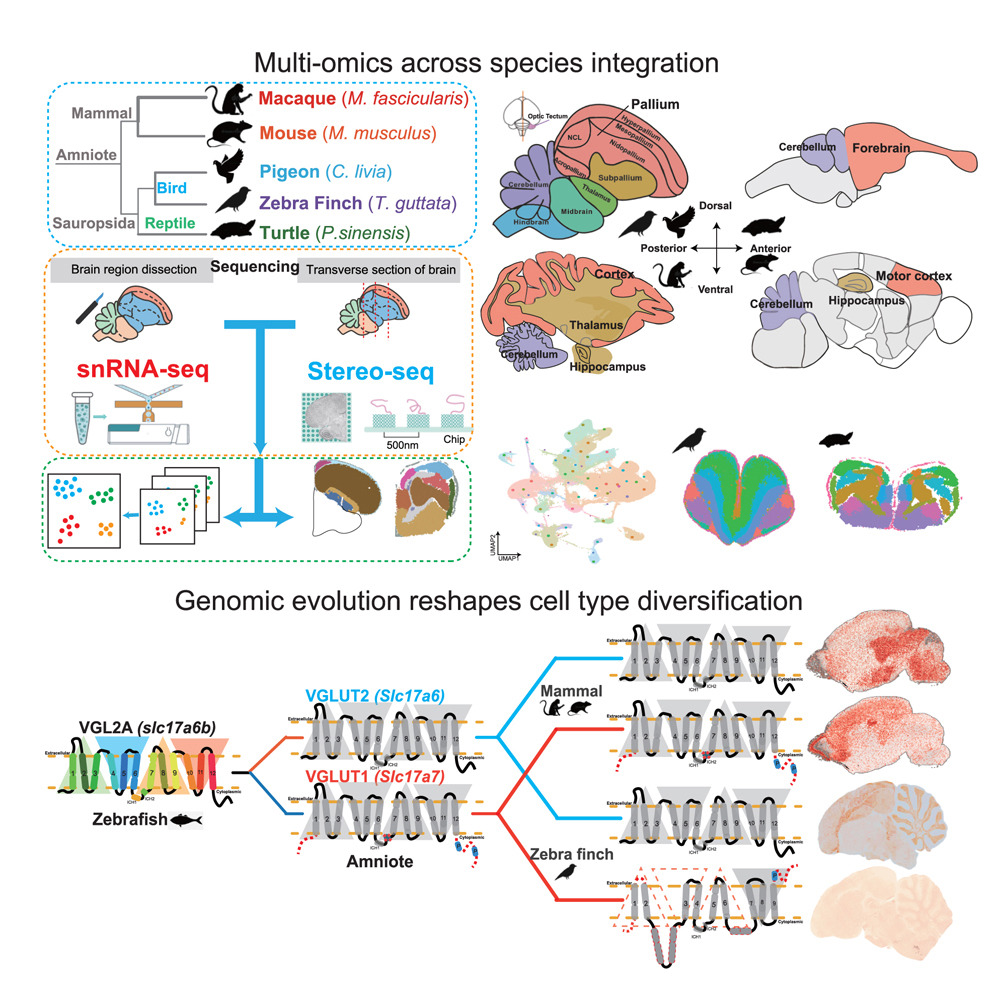 Genomic evolution reshapes cell-type diversification in the amniote brainDuoyuan Chen† , Zhenkun Zhuang† , Maolin Huang† , Yunqi Huang† , Yuting Yan , Yanru Zhang , Youning Lin , Xiaoying Jin , Yuanmei Wang , Jinfeng Huang , Wenbo Xu , Jingfang Pan , Hong Wang , Fubaoqian Huang , Kuo Liao , Mengnan Cheng , Zhiyong Zhu , Yinqi Bai , Zhiwei Niu , Ze Zhang , Ya Xiang , Xiaofeng Wei , Tao Yang , Tao Zeng , Yuliang Dong , Ying Lei , Yangang Sun , Jian Wang , Huanming Yang , Yidi Sun , Gang Cao , Muming Poo , Longqi Liu , Robert K. Naumann* , Chun Xu* , Zhenlong Wang* , Xun Xu* , and Shiping Liu*Developmental Cell, May 2025
Genomic evolution reshapes cell-type diversification in the amniote brainDuoyuan Chen† , Zhenkun Zhuang† , Maolin Huang† , Yunqi Huang† , Yuting Yan , Yanru Zhang , Youning Lin , Xiaoying Jin , Yuanmei Wang , Jinfeng Huang , Wenbo Xu , Jingfang Pan , Hong Wang , Fubaoqian Huang , Kuo Liao , Mengnan Cheng , Zhiyong Zhu , Yinqi Bai , Zhiwei Niu , Ze Zhang , Ya Xiang , Xiaofeng Wei , Tao Yang , Tao Zeng , Yuliang Dong , Ying Lei , Yangang Sun , Jian Wang , Huanming Yang , Yidi Sun , Gang Cao , Muming Poo , Longqi Liu , Robert K. Naumann* , Chun Xu* , Zhenlong Wang* , Xun Xu* , and Shiping Liu*Developmental Cell, May 2025Over 320 million years of evolution, amniotes have developed complex brains and cognition through largely unexplored genetic and gene expression mechanisms. We created a comprehensive single-cell atlas of over 1.3 million cells from the telencephalon and cerebellum of turtles, zebra finches, pigeons, mice, and macaques, employing single-cell resolution spatial transcriptomics to validate gene expression patterns across species. Our study identifies significant species-specific variations in cell types, highlighting their conservation and diversification in evolution. We found pronounced differences in telencephalon excitatory neurons (EXs) and cerebellar cell types between birds and mammals. Birds predominantly express SLC17A6 in EX, whereas mammals express SLC17A7 in the neocortex and SLC17A6 elsewhere, possibly due to loss of function of SLC17A7 in birds. Additionally, we identified a bird-specific Purkinje cell subtype (SVIL+), implicating the lysine-specific demethylase 11 (LSD1)/KDM1A pathway in learning and circadian rhythms and containing numerous positively selected genes, which suggests an evolutionary optimization of cerebellar functions for ecological and behavioral adaptation. Our findings elucidate the complex interplay between genetic evolution and environmental adaptation, underscoring the role of genetic diversification in the development of specialized cell types across amniotes.
@article{10.1016/j.devcel.2025.04.014, author = {Chen†, Duoyuan and Zhuang†, Zhenkun and Huang†, Maolin and Huang†, Yunqi and Yan, Yuting and Zhang, Yanru and Lin, Youning and Jin, Xiaoying and Wang, Yuanmei and Huang, Jinfeng and Xu, Wenbo and Pan, Jingfang and Wang, Hong and Huang, Fubaoqian and Liao, Kuo and Cheng, Mengnan and Zhu, Zhiyong and Bai, Yinqi and Niu, Zhiwei and Zhang, Ze and Xiang, Ya and Wei, Xiaofeng and Yang, Tao and Zeng, Tao and Dong, Yuliang and Lei, Ying and Sun, Yangang and Wang, Jian and Yang, Huanming and Sun, Yidi and Cao, Gang and Poo, Muming and Liu, Longqi and Naumann*, Robert K. and Xu*, Chun and Wang*, Zhenlong and Xu*, Xun and Liu*, Shiping}, title = {Genomic evolution reshapes cell-type diversification in the amniote brain}, journal = {Developmental Cell}, year = {2025}, month = may, day = {13}, pages = {1-60}, volume = {60}, number = {}, doi = {10.1016/j.devcel.2025.04.014}, url = {https://www.sciencedirect.com/science/article/pii/S1534580725002527}, eprint = {https://www.sciencedirect.com/science/article/pii/S1534580725002527}, } -
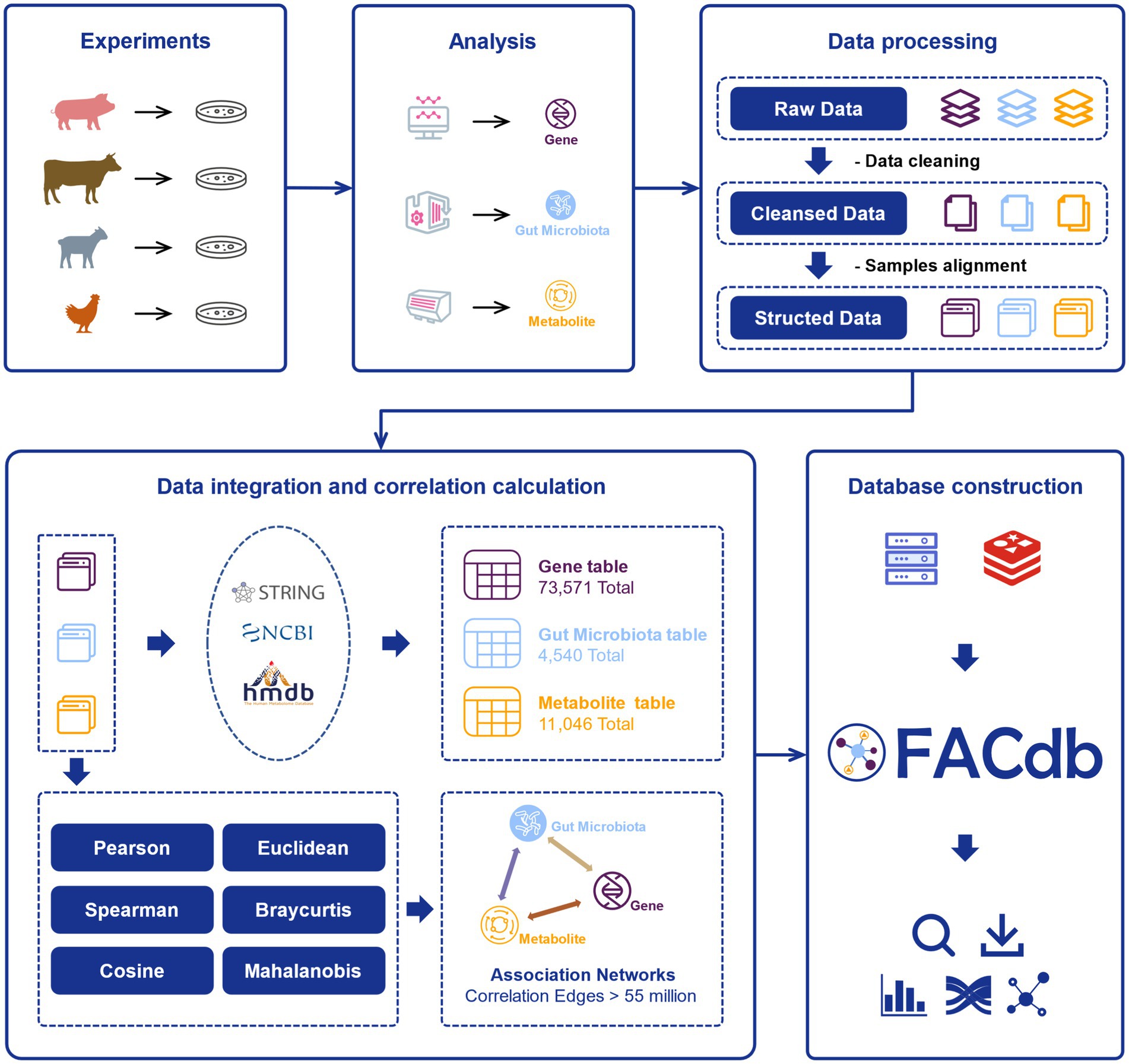 FACdb: a comprehensive resource for genes, gut microbiota, and metabolites in farm animalsZe Zhang† , Yang Li† , Di Zhang† , Shuai Chen* , Sien Lu , Kang Wang , Miao Zhou , Zehe Song , Qingcui Li* , Jie Yin* , and Xiaoping Liu*Frontiers in Microbiology, Mar 2025
FACdb: a comprehensive resource for genes, gut microbiota, and metabolites in farm animalsZe Zhang† , Yang Li† , Di Zhang† , Shuai Chen* , Sien Lu , Kang Wang , Miao Zhou , Zehe Song , Qingcui Li* , Jie Yin* , and Xiaoping Liu*Frontiers in Microbiology, Mar 2025Farm animals, including livestock and poultry, play essential economic, social, and cultural roles and are indispensable in human welfare. Farm Animal Connectome database (FACdb) is a comprehensive resource that includes the association networks among gene expression, gut microbiota, and metabolites in farm animals. Although some databases present the relationship between gut microbes, metabolites, and gene expression, these databases are limited to human and mouse species, with limited data for farm animals. In this database, we calculate the associations and summarize the connections among gene expression, gut microbiota, and metabolites in farm animals using six correlation or distance calculation (including Pearson, Spearman, Cosine, Euclidean, Bray–Curtis, and Mahalanobis). FACdb contains over 55 million potential interactions of 73,571 genes, 11,046 gut microbiota, and 4,540 metabolites. It provides an easy-to-use interface for browsing and searching the association information. Additionally, FACdb offers interactive visualization tools to effectively investigate the relationship among the genes, gut microbiota, and metabolites in farm animals. Overall, FACdb is a valuable resource for understanding interactions among gut microbiota, metabolites, and gene expression. It contributes to the further utilization of microbes in animal products and welfare promotion. Compared to mice, pigs or other farm animals share more similarities with humans in molecular, cellular, and organ-level responses, indicating that our database may offer new insights into the relationship among gut microbiota, metabolites, and gene expression in humans.
@article{10.3389/fmicb.2025.1557285, author = {Zhang†, Ze and Li†, Yang and Zhang†, Di and Chen*, Shuai and Lu, Sien and Wang, Kang and Zhou, Miao and Song, Zehe and Li*, Qingcui and Yin*, Jie and Liu*, Xiaoping}, title = {FACdb: a comprehensive resource for genes, gut microbiota, and metabolites in farm animals}, journal = {Frontiers in Microbiology}, year = {2025}, month = mar, day = {21}, pages = {}, volume = {16}, number = {}, doi = {10.3389/fmicb.2025.1557285}, url = {https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1557285/full}, eprint = {https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1557285/full}, }
2024
-
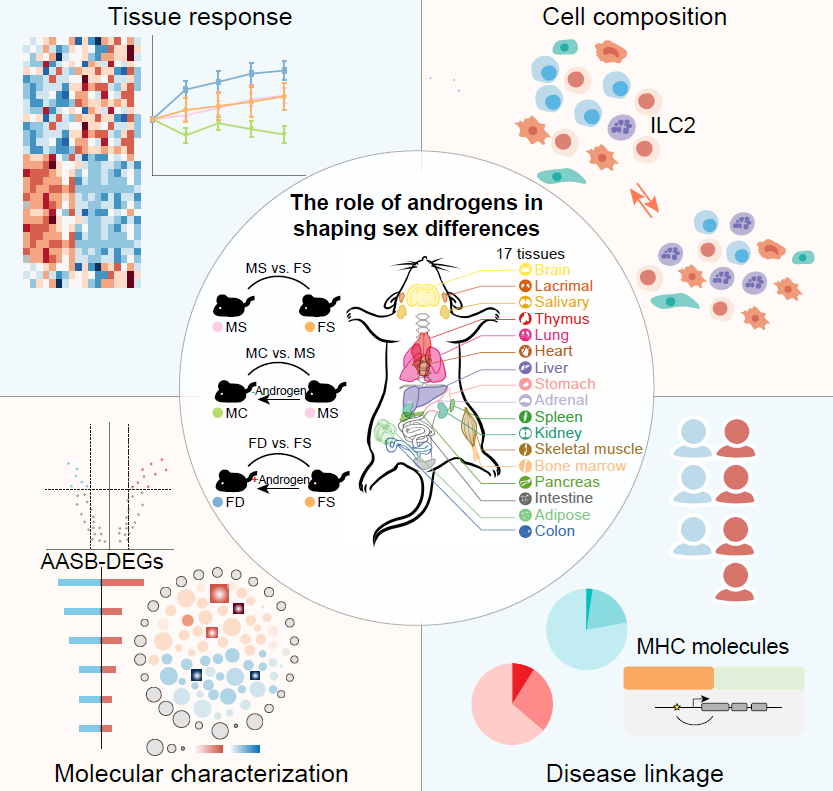 Sex differences orchestrated by androgens at single-cell resolutionFei Li† , Xudong Xing† , Qiqi Jin† , Xiang-Ming Wang† , Pengfei Dai† , Ming Han† , Huili Shi , Ze Zhang , Xianlong Shao , Yunyi Peng , Yiqin Zhu , Jiayi Xu , Dan Li , Yu Chen , Wei Wu , Qiao Wang , Chen Yu* , Luonan Chen* , Fan Bai* , and Dong Gao*Nature, Apr 2024
Sex differences orchestrated by androgens at single-cell resolutionFei Li† , Xudong Xing† , Qiqi Jin† , Xiang-Ming Wang† , Pengfei Dai† , Ming Han† , Huili Shi , Ze Zhang , Xianlong Shao , Yunyi Peng , Yiqin Zhu , Jiayi Xu , Dan Li , Yu Chen , Wei Wu , Qiao Wang , Chen Yu* , Luonan Chen* , Fan Bai* , and Dong Gao*Nature, Apr 2024Sex differences in mammalian complex traits are prevalent and are intimately associated with androgens1-7. However, a molecular and cellular profile of sex differences and their modulation by androgens is still lacking. Here we constructed a high-dimensional single-cell transcriptomic atlas comprising over 2.3 million cells from 17 tissues in Mus musculus and explored the effects of sex and androgens on the molecular programs and cellular populations. In particular, we found that sex-biased immune gene expression and immune cell populations, such as group 2 innate lymphoid cells, were modulated by androgens. Integration with the UK Biobank dataset revealed potential cellular targets and risk gene enrichment in antigen presentation for sex-biased diseases. This study lays the groundwork for understanding the sex differences orchestrated by androgens and provides important evidence for targeting the androgen pathway as a broad therapeutic strategy for sex-biased diseases.
@article{doi:10.1038/s41586-024-07291-6, author = {Li†, Fei and Xing†, Xudong and Jin†, Qiqi and Wang†, Xiang-Ming and Dai†, Pengfei and Han†, Ming and Shi, Huili and Zhang, Ze and Shao, Xianlong and Peng, Yunyi and Zhu, Yiqin and Xu, Jiayi and Li, Dan and Chen, Yu and Wu, Wei and Wang, Qiao and Yu*, Chen and Chen*, Luonan and Bai*, Fan and Gao*, Dong}, title = {Sex differences orchestrated by androgens at single-cell resolution}, journal = {Nature}, volume = {}, number = {}, pages = {1-8}, year = {2024}, month = apr, day = {10}, doi = {10.1038/s41586-024-07291-6}, url = {https://www.nature.com/articles/s41586-024-07291-6}, eprint = {https://www.nature.com/articles/s41586-024-07291-6}, }
2023
-
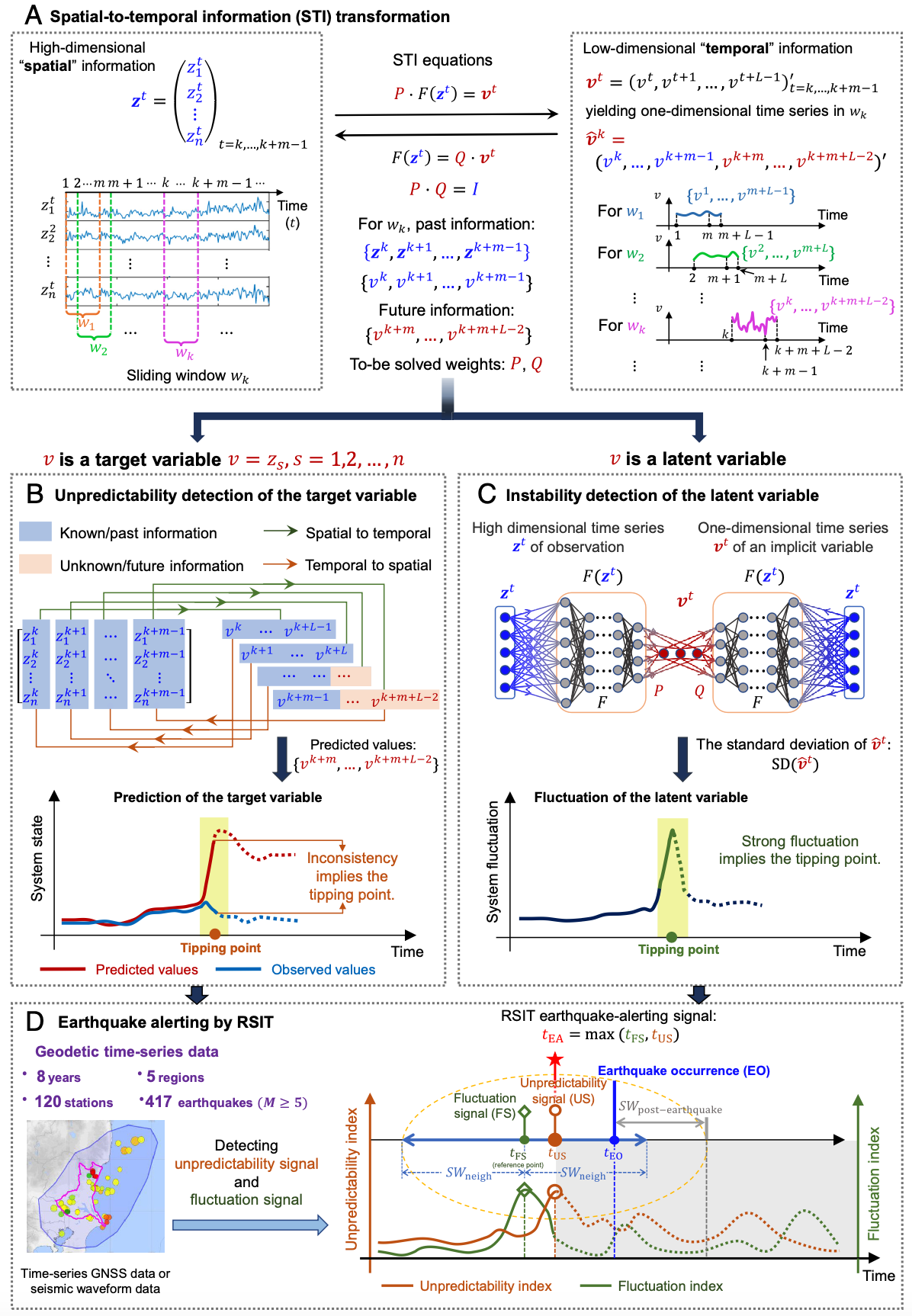 Earthquake alerting based on spatial geodetic data by spatiotemporal information transformation learningYuyan Tong† , Renhao Hong† , Ze Zhang , Kazuyuki Aihara , Pei Chen* , Rui Liu* , and Luonan Chen*Proceedings of the National Academy of Sciences, Sep 2023
Earthquake alerting based on spatial geodetic data by spatiotemporal information transformation learningYuyan Tong† , Renhao Hong† , Ze Zhang , Kazuyuki Aihara , Pei Chen* , Rui Liu* , and Luonan Chen*Proceedings of the National Academy of Sciences, Sep 2023Alerting for imminent earthquakes is particularly challenging due to the high nonlinearity and nonstationarity of geodynamical phenomena. In this study, based on spatiotemporal information (STI) transformation for high-dimensional real-time data, we developed a model-free framework, i.e., real-time spatiotemporal information transformation learning (RSIT), for extending the nonlinear and nonstationary time series. Specifically, by transforming high-dimensional information of the global navigation satellite system into one-dimensional dynamics via the STI strategy, RSIT efficiently utilizes two criteria of the transformed one-dimensional dynamics, i.e., unpredictability and instability. Such two criteria contemporaneously signal a potential critical transition of the geodynamical system, thereby providing early-warning signals of possible upcoming earthquakes. RSIT explores both the spatial and temporal dynamics of real-world data on the basis of a solid theoretical background in nonlinear dynamics and delay-embedding theory. The effectiveness of RSIT was demonstrated on geodynamical data of recent earthquakes from a number of regions across at least 4 y and through further comparison with existing methods.
@article{doi:10.1073/pnas.2302275120, author = {Tong†, Yuyan and Hong†, Renhao and Zhang, Ze and Aihara, Kazuyuki and Chen*, Pei and Liu*, Rui and Chen*, Luonan}, title = {Earthquake alerting based on spatial geodetic data by spatiotemporal information transformation learning}, journal = {Proceedings of the National Academy of Sciences}, volume = {120}, number = {37}, pages = {e2302275120}, year = {2023}, month = sep, day = {05}, doi = {10.1073/pnas.2302275120}, url = {https://www.pnas.org/doi/abs/10.1073/pnas.2302275120}, eprint = {https://www.pnas.org/doi/pdf/10.1073/pnas.2302275120}, } -
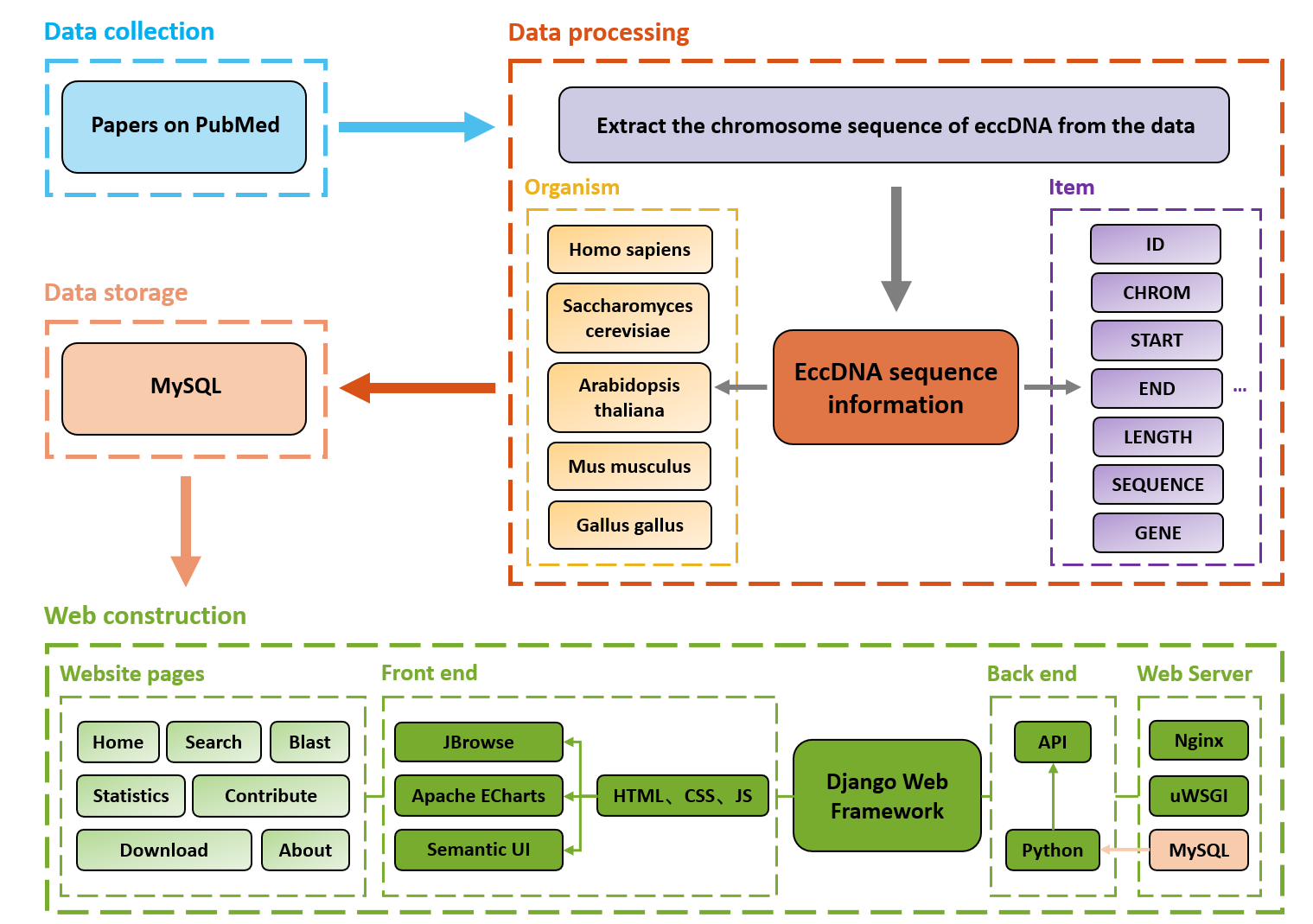 TeCD: The eccDNA Collection Database for extrachromosomal circular DNAJing Guo† , Ze Zhang† , Qingcui Li , Xiao Chang* , and Xiaoping Liu*BMC Genomics, Jan 2023
TeCD: The eccDNA Collection Database for extrachromosomal circular DNAJing Guo† , Ze Zhang† , Qingcui Li , Xiao Chang* , and Xiaoping Liu*BMC Genomics, Jan 2023Background Extrachromosomal circular DNA (eccDNA) is a kind of DNA that widely exists in eukaryotic cells. Studies in recent years have shown that eccDNA is often enriched during tumors and aging, and participates in the development of cell physiological activities in a special way, so people have paid more and more attention to the eccDNA, and it has also become a critical new topic in modern biological research. Description We built a database to collect eccDNA, including animals, plants and fungi, and provide researchers with an eccDNA retrieval platform. The collected eccDNAs were processed in a uniform format and classified according to the species to which it belongs and the chromosome of the source. Each eccDNA record contained sequence length, start and end sites on the corresponding chromosome, order of the bases, genomic elements such as genes and transposons, and other information in the respective sequencing experiment. All the data were stored into the TeCD (The eccDNA Collection Database) and the BLAST (Basic Local Alignment Search Tool) sequence alignment function was also added into the database for analyzing the potential eccDNA sequences. Conclusion We built TeCD, a platform for users to search and obtain eccDNA data, and analyzed the possible potential functions of eccDNA. These findings may provide a basis and direction for researchers to further explore the biological significance of eccDNA in the future.
@article{doi:10.1186/s12864-023-09135-5, author = {Guo†, Jing and Zhang†, Ze and Li, Qingcui and Chang*, Xiao and Liu*, Xiaoping}, title = {TeCD: The eccDNA Collection Database for extrachromosomal circular DNA}, journal = {BMC Genomics}, volume = {24}, number = {1}, pages = {47}, year = {2023}, month = jan, day = {27}, doi = {10.1186/s12864-023-09135-5}, url = {https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-023-09135-5}, eprint = {https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-023-09135-5}, }